Special kinds of BLASTs
In addition to the standard BLAST algorithms (BLASTn, BLASTp, BLASTx, tBLASTn, tBLASTx), there are several special kinds of BLASTs that have been developed to address specific needs in sequence analysis. Here are a few examples:
Advertisements

- PSI-BLAST (Position-Specific Iterated BLAST): PSI-BLAST is an iterative version of BLASTp that aims to improve the detection of distantly related protein sequences. It builds a position-specific scoring matrix (PSSM) based on the alignments found in previous iterations, allowing for the identification of more divergent homologs.

- PHI-BLAST (Pattern-Hit Initiated BLAST): PHI-BLAST is used for identifying and aligning protein sequences that contain specific patterns or motifs. It starts with a pattern search against a protein database and then extends the search using a BLAST-like algorithm.
- DELTA-BLAST: DELTA-BLAST is a tool that combines the advantages of PSI-BLAST and HMMER. It performs a search using PSI-BLAST and then uses the identified homologs to construct a position-specific scoring matrix (PSSM), which is further used to perform a search with HMMER against a custom database.
- Reverse-BLAST: Reverse-BLAST is used to find potential nucleotide or protein sources for a given sequence. It takes a query sequence and searches for its potential sources by comparing it against a database of known sequences.
- Short-Read BLAST (SR-BLAST): SR-BLAST is designed to handle the analysis of short DNA sequence reads generated by high-throughput sequencing technologies. It allows for efficient and accurate alignment of short reads against a reference database, enabling tasks such as read mapping and variant calling.
These specialized BLAST variants cater to specific research needs and provide enhanced capabilities for analyzing different types of sequences or addressing specific sequence analysis challenges.
Characteristics of BLAST
BLAST (Basic Local Alignment Search Tool) possesses a number of essential characteristics that contribute to its efficacy and pervasive application in sequence analysis. Here are some distinguishing features of BLAST:
Advertisements
- Speed and Efficiency: BLAST is designed to perform sequence similarity searches quickly and efficiently. It utilizes heuristic algorithms and indexing techniques to expedite the identification of local alignments, making it suitable for searching large sequence databases in a reasonable amount of time.
- Sensitivity and Specificity: In sequence comparisons, BLAST establishes a balance between sensitivity and specificity. It seeks to identify meaningful correlations while minimizing false positives. BLAST provides a measure of the statistical significance of the identified matches by utilizing scoring matrices and statistical measures such as E-values.
- Focus on Local Alignments: BLAST focuses on identifying local rather than global alignments. It identifies shorter regions of substantial similarity, known as high-scoring segment pairs (HSPs), which enables efficient identification of conserved regions even in sequences with divergent characteristics.
- Iterative Method: Some BLAST variants, such as PSI-BLAST, employ an iterative method. They conduct multiple cycles of searching and alignment to refine the query and database sequences progressively. This iterative procedure facilitates the detection of more distant homologs and increases sensitivity.
- Flexibility: BLAST is versatile and can be applied to numerous categories of biological sequences, such as DNA, RNA, and proteins. Different BLAST variants are tailored to specific sequence types and search criteria, allowing for versatility in sequence analysis duties.
- User-Friendly Interface: BLAST tools typically feature user-friendly interfaces that enable researchers to readily input query sequences, select databases, and configure search parameters. This accessibility enables users with differing degrees of bioinformatics knowledge to conduct efficient sequence similarity searches.

- Extensive Database Compatibility: BLAST is compatible with a vast array of sequence databases, including public databases such as GenBank, UniProt, and the NCBI’s non-redundant (nr) database. This compatibility enables researchers to compare their sequences to exhaustive collections of previously identified sequences.
- Community Support and Updates: BLAST has a sizable user community, which has aided in its ongoing development and updates. Regular updates and issue fixes ensure that BLAST remains a trustworthy and current sequence analysis tool.
How BLAST Works
- The BLAST algorithm is a heuristic program, which means it uses intelligent shortcuts to perform the search more quickly.

- BLAST performs “local” alignments. Functional domains are frequently repeated within the same protein as well as across proteins from different species in the vast majority of proteins.
- The BLAST algorithm is optimized to identify these domains or shorter sequence-similar segments. Local alignment also allows an mRNA to be aligned with a fragment of genomic DNA, which is frequently necessary for genome assembly and analysis.
- If BLAST initially attempted to align two sequences along their entire lengths (known as a global alignment), fewer similarities would be detected, particularly in terms of domains and motifs.
- When a query is submitted through one of the BLAST Web pages, the sequence, along with any other input information such as the database to be searched, word size, expected value, etc., is supplied to the algorithm on the BLAST server.
- BLAST operates by first creating a look-up table of all the “words” (brief subsequences, which for proteins have a default length of three letters) and “neighboring words,” i.e., words in the query sequence that are similar to the query words.
- The sequence database is then searched for these “hot spots” When a match is found, it is utilized to generate gap-free and gapped extensions of the “word.” Directly searching GenBank flatfiles (or any subset of GenBank flatfiles) is not supported by BLAST.
- Sequences are instead added to BLAST databases. Each entry is divided into two files, one containing only the header information and the other containing only the sequence information.
- These are the data utilized by the algorithm. If BLAST is to be executed in “stand-alone” mode, the data file may contain local, private data, downloaded NCBI BLAST databases, or a combination of both.
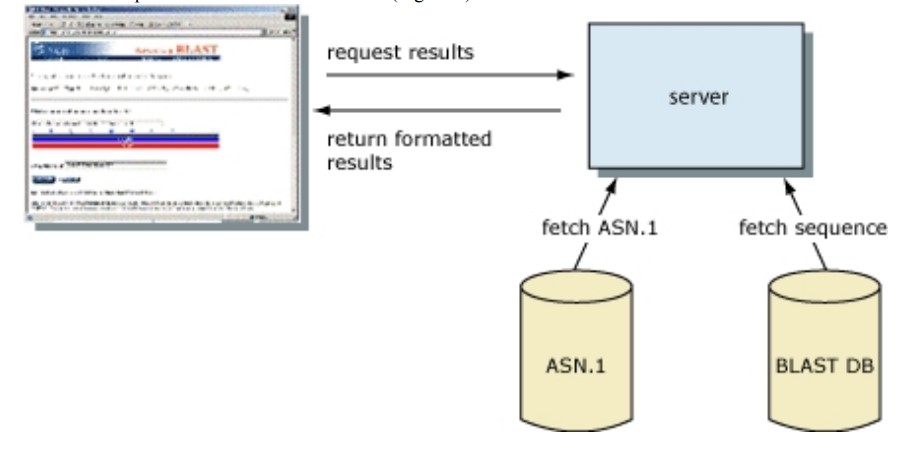
- After the algorithm has searched for and maximally extended all possible “words” from the query sequence, it assembles the best alignment for each query–sequence pair and writes this information to a SeqAlign data structure. The SeqAlign structure does not contain sequence information; instead, it references the sequences in the BLAST database.
- The BLAST Formatter, which resides on the BLAST server, can utilize the information in the SeqAlign to retrieve and display similar sequences in a variety of ways. Therefore, once a query has been executed, the results can be reformatted without rerunning the search. This is made feasible by the QBLAST system.

Comments
Post a Comment