THE CELL CYCLE AND MITOSIS
INTRODUCTION
TOPIC 8.1: REGULATING THE CELL CYCLE: CHECKPOINT CONTROL
Learning Goals
- Review the stages of the cell cycle, including the checkpoints, and identify the key features of each stage.
- Describe how specific protein modifications (e.g., phosphorylation and ubiquitination) result in activation/deactivation of cyclin-CDK complexes to regulate cell cycle checkpoints.
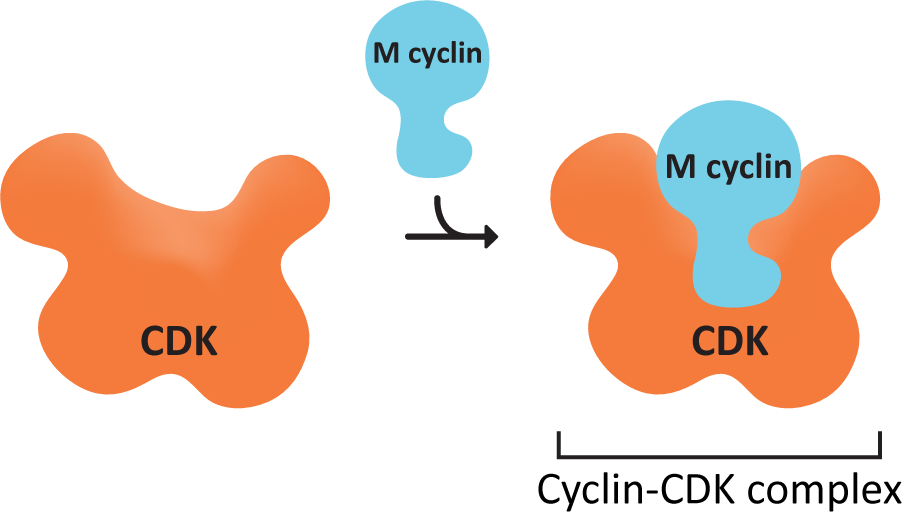
- Explain how the activation of the cyclin-CDK complexes results in the start of the next phase of the cell cycle. Use examples from both M- and S-cyclin-CDK complexes to explain this.
- Detail how fluorescence-activated cell sorting (FACS) can be used to identify the stage of the cell cycle for a population of cells.
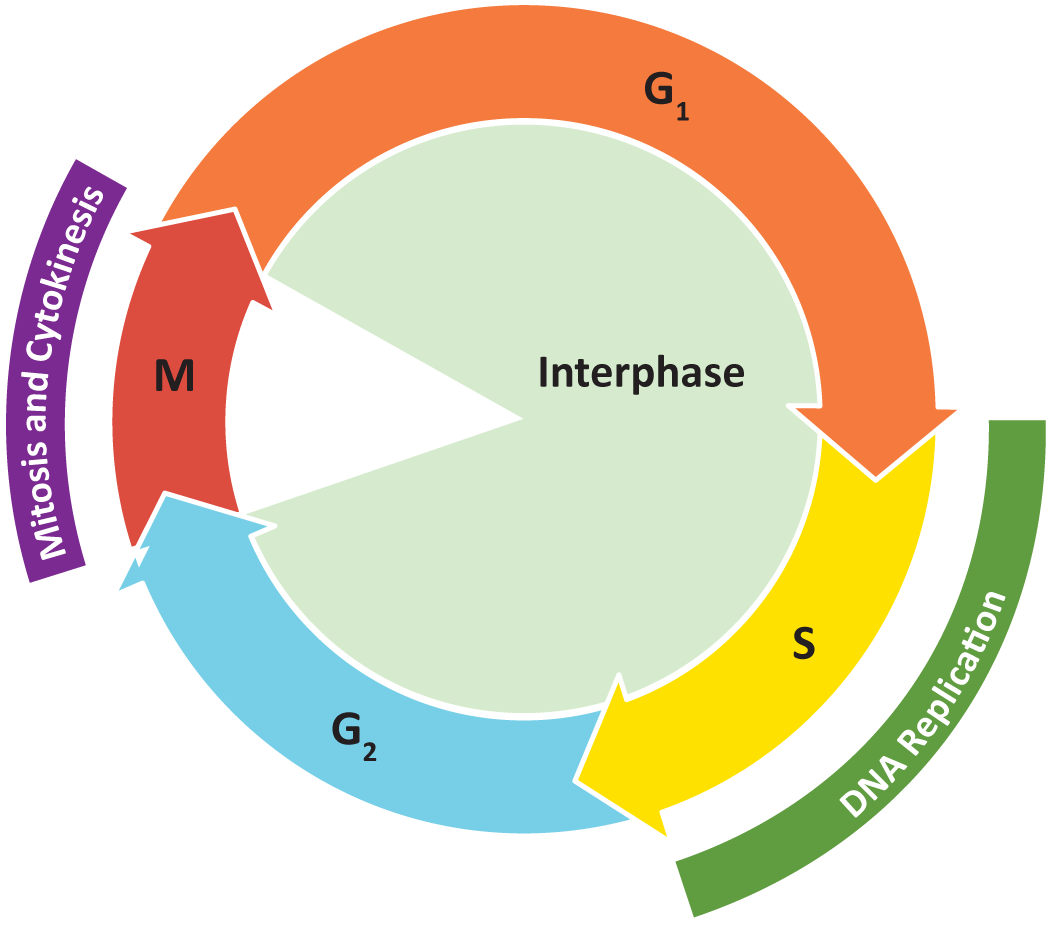
- G1 (gap or growth 1) phase: This is the “gap” between the end of cytokinesis and the start of DNA synthesis. A lot of the work of this phase involves cell growth so that it can support itself and also have the resources it needs for the next phase.
- S (synthesis) phase: This phase is defined by the initiation and termination of DNA synthesis.
- G2 (gap or growth 2) phase: This second “gap” phase lasts from the end of DNA synthesis to the onset of mitosis. The cell continues to grow but also prepares for what’s to come in the next phase.
- M (mitosis) phase: This is the phase in which cell division occurs.
- discrete events, such as DNA synthesis and mitosis, do not occur before the cell is ready, and they occur in the right order
- the cell pauses DNA replication or mitosis when errors are identified and repairs are attempted
- only cells that should divide are allowed to do so
- Some cells must undergo terminal differentiation, stall temporarily or permanently, or even go through programmed cell death (i.e., apoptosis). These are all cell cycle decisions.
- growth is coordinated with cell division so that the size of cells is maintained over the many cellular generations
- Some cells are designed to get bigger or smaller over several cell cycles. Some even divide asymmetrically so that one large and one small daughter cell is produced. All of these will require precise cell cycle control.
- Proper nutrients (carbon source, energy source, inorganic phosphate, nitrogen, vitamins, etc.) must be present at specific concentrations.
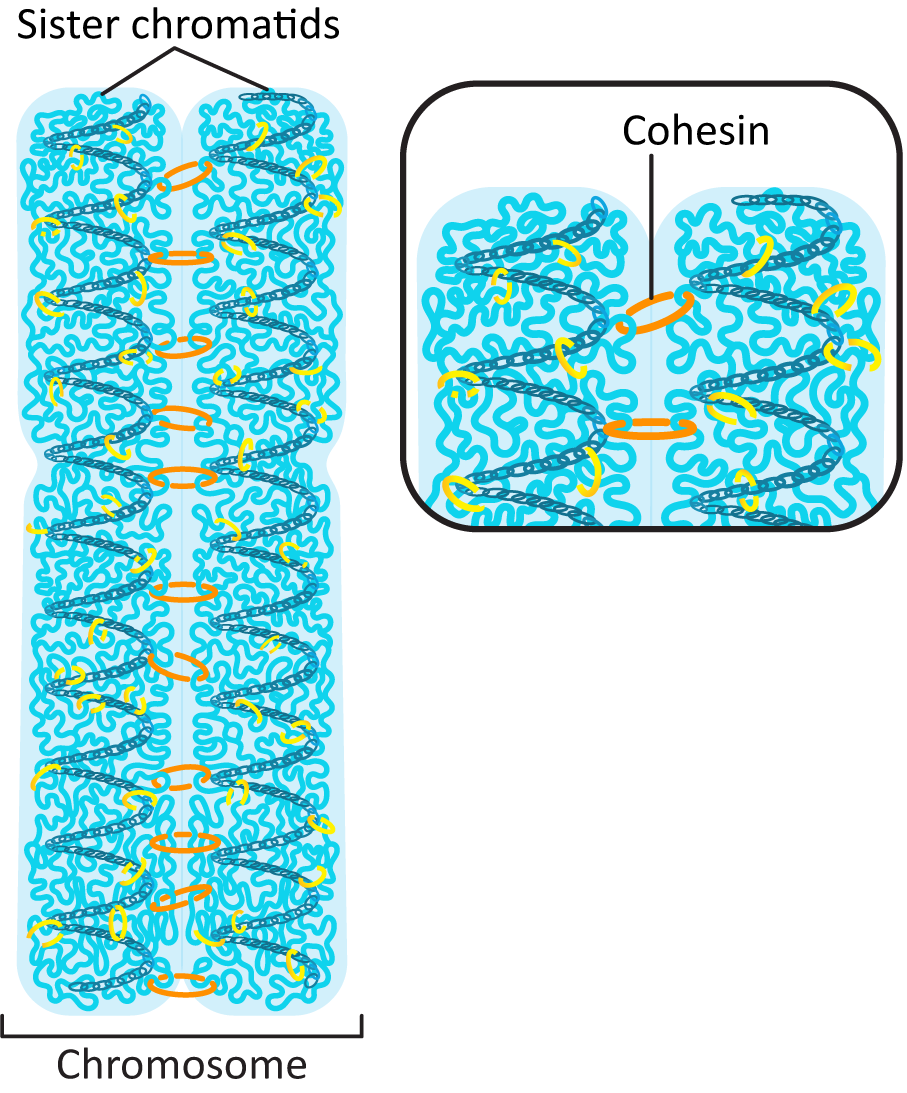
- Sister chromatid separation (from the previous mitosis) must be complete.
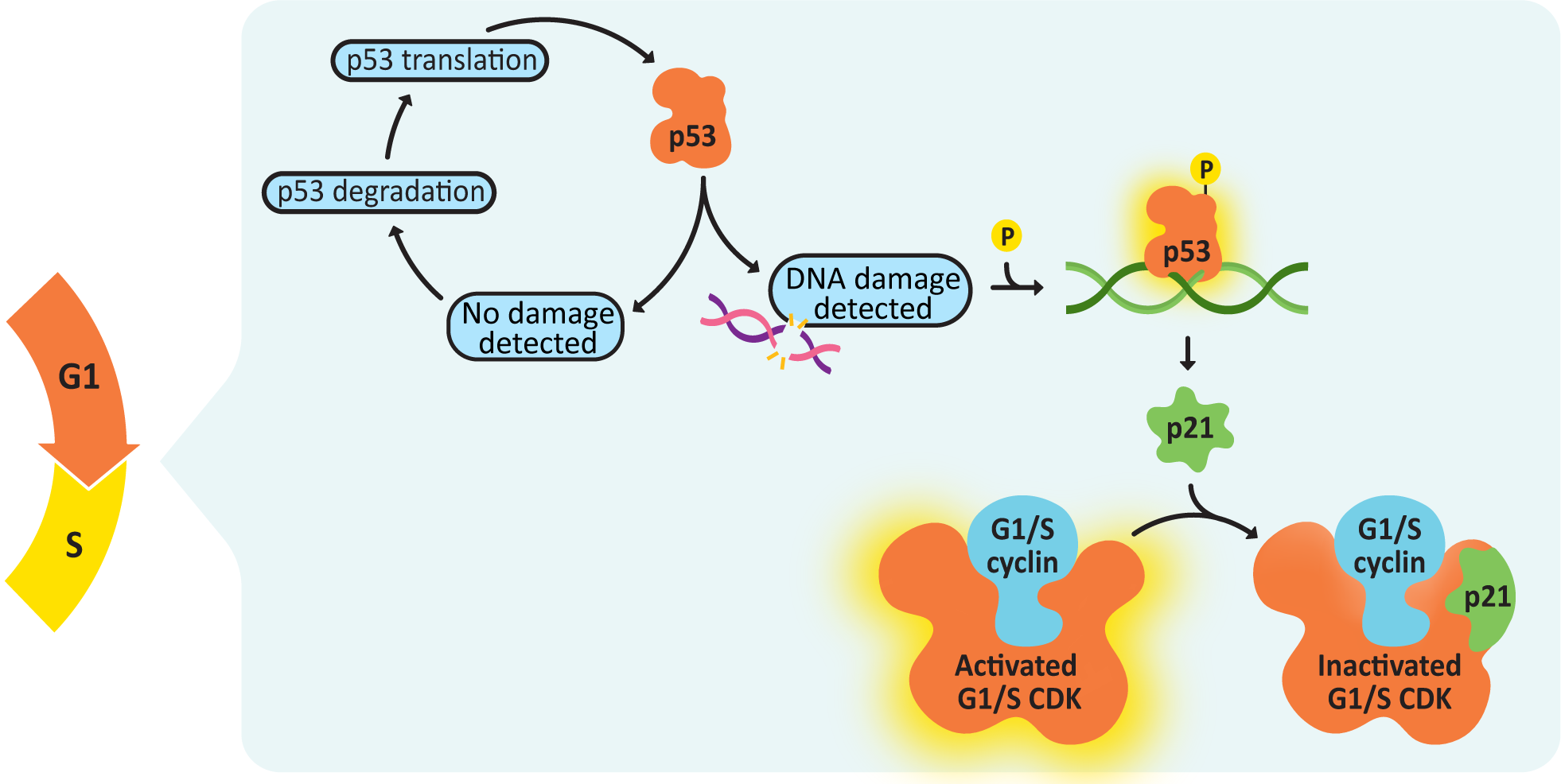
- There must be no detectable DNA damage.
- The cell must have reached a critical threshold size.
- Stem cells for specific tissues will enter G0 for short periods of time until replacement cells are required. This allows tissues to grow to a certain size and then stop growing and maintain a relatively stable size and distribution. Your blood cell system (called the hematopoietic system) does this. New blood cells are grown only when specific cell types are needed or cells are lost through injury.
- Some cell types undergo terminal differentiation, which means that when these cells reach maturity, they do not need to go through mitosis anymore. Good examples of this are muscle cells (multiple muscle cells fuse together at maturity to produce multinucleate muscle fibers), neurons (with their extremely long axons), and osteocytes (bone cells, which are intricately embedded in the calcified matrix of the bone).
- DNA replication must be complete and accurate. No DNA damage can be detected (through a robust biochemical surveillance system) or this checkpoint cannot be passed. This is the most important factor for passing the G2/M checkpoint.
- The cell must also have reached a certain minimum size so that it is big enough that, when split in two, the two daughter cells will also be large enough to survive.
- The active agent that promotes the next stage of the cell cycle is in the cytosol.
- Control of DNA synthesis and mitosis is positive—that is, the active agent promotes mitosis in the recipient cell.
- Cells can be advanced into the next stage before they planned it by adding the appropriate factors to their cytosol.
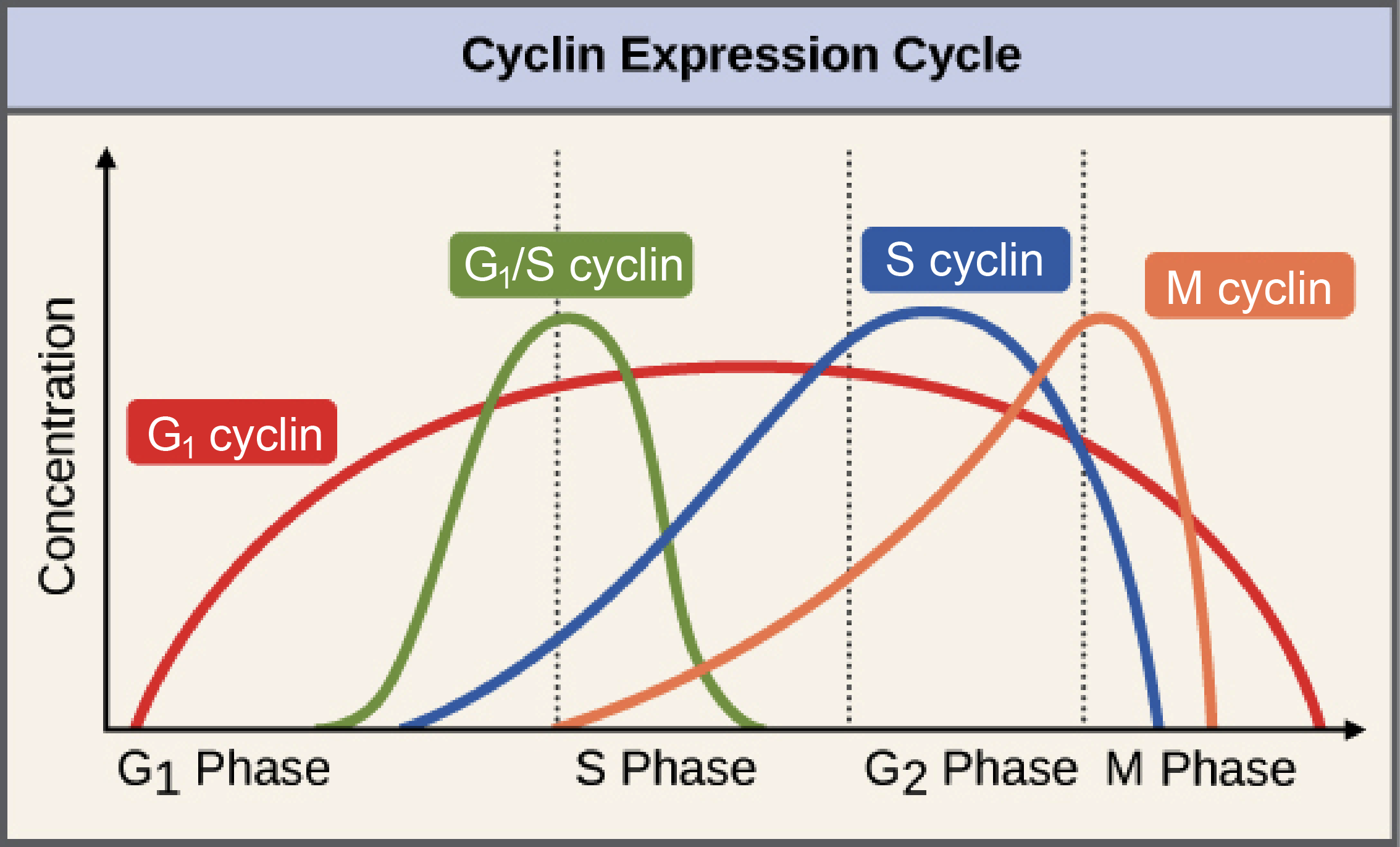
| MITOTIC CYCLIN CLASSES | |
|---|---|
| CYCLIN CLASS | FUNCTION |
| G1 cyclins | Unique cyclins that are thought to help the cell respond to external signals to leave G0 and initiate cell division. We won’t discuss these any further in this book. |
| G1/S cyclins | Cyclins that control the G1/S checkpoint and control the transition from G1 to S phase. |
| S cyclins | Cyclins that activate at the start of S phase (by the G1/S cyclins) and directly induce replication of DNA. The concentration of these cyclins remains high right through to M phase. |
| M cyclins | Cyclins that control the G2/M checkpoint. They remain active in the first half of mitosis until their destruction is signaled by the anaphase-promoting complex (APC). |
| THE EFFECTS OF MUTATIONS IN WEE1 AND CDC25 ON YEAST CELL GROWTH AND DIVISION | ||
|---|---|---|
| GENE | GAIN-OF-FUNCTION MUTATION* | LOSS-OF-FUNCTION MUTATION** |
| WEE1—inhibitory kinase | Cells divide later at a larger-than-normal size | Cells divide early at a smaller-than-normal size (i.e., they’re “wee”!) |
| CDC25—activating phosphatase | Cells divide early at a smaller-than-normal size | Cells divide later at a larger-than-normal size |
| *i.e., too much protein produced or produced all of the time without regulation **i.e., no protein produced | ||
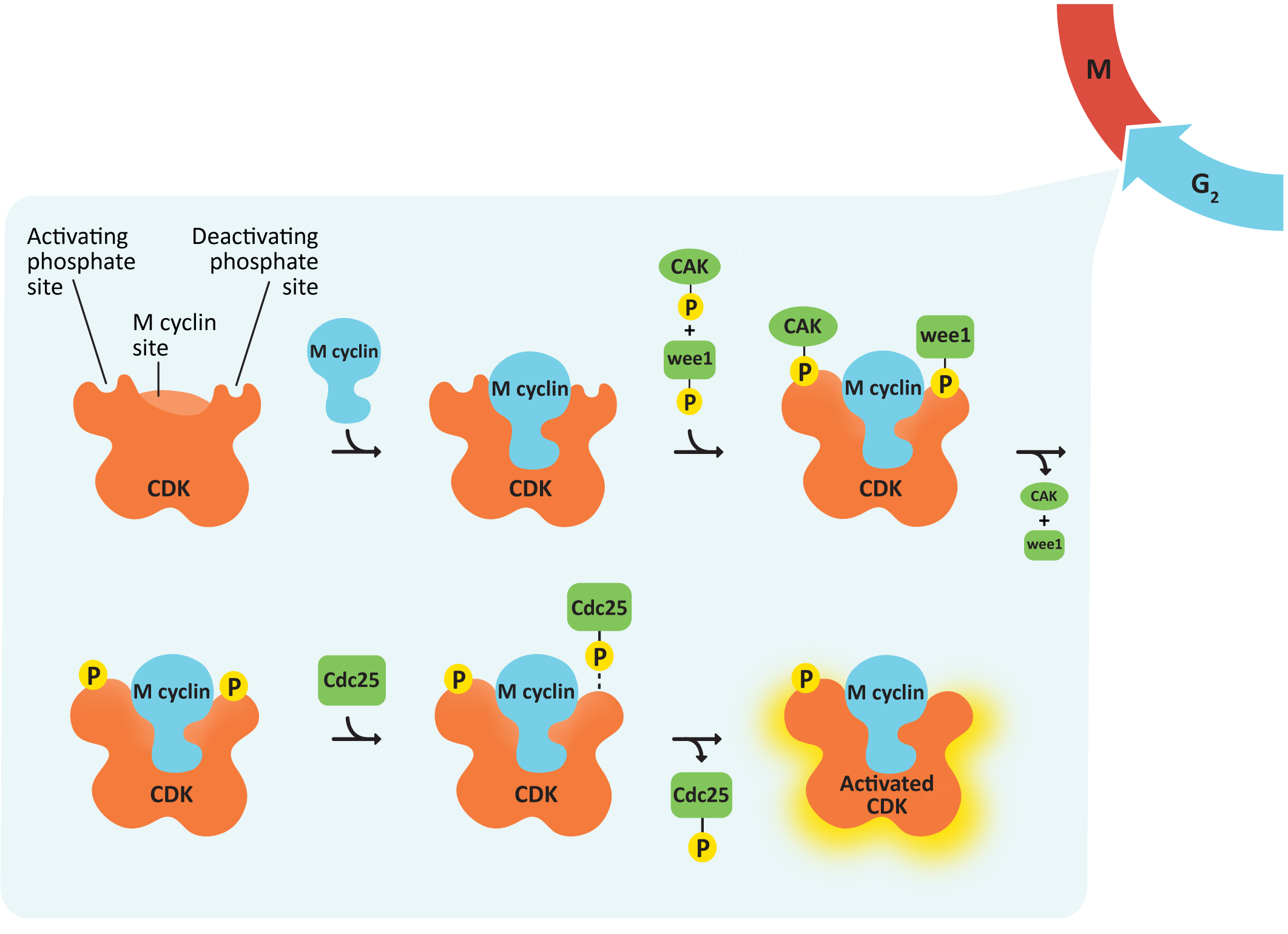
Control of the G1/S Checkpoint and S Phase Progression
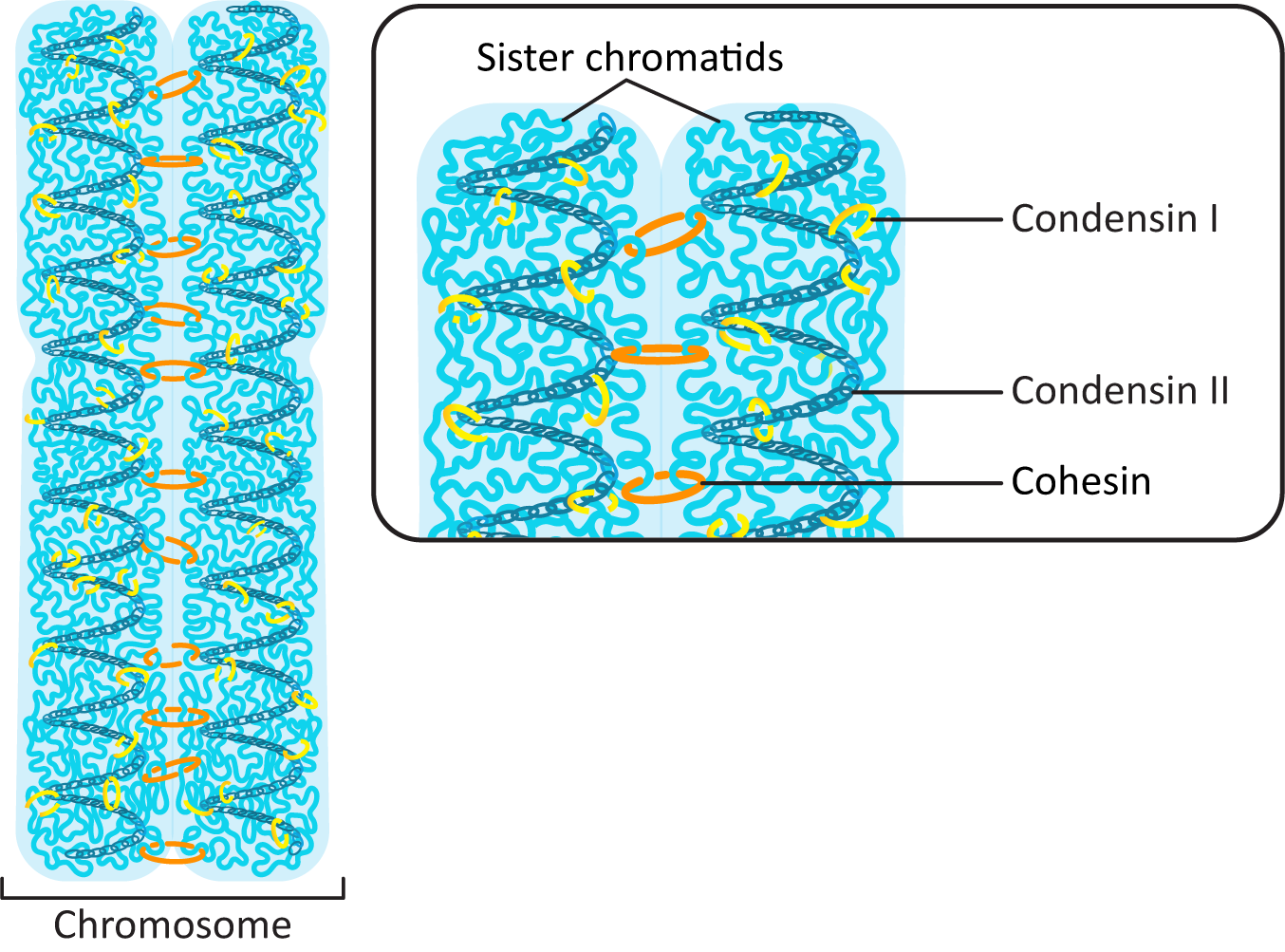
- Histone H1—Phosphorylating H1 leads to changes in chromatin configuration and, in conjunction with other proteins, leads to the tighter packing of chromatin required for mitosis.
- Condensins—These are a class of DNA-binding proteins that bind to chromatin to help with higher-order chromosome condensation. They work to loop up the chromatin fiber into the tightly coiled mitotic chromosome.
- Nuclear lamins—Phosphorylated lamins have a lower affinity for each other, and as such, the nuclear lamina falls apart. Disassembly of nuclear lamina results in the breakup of the nuclear envelope. (We explored this in detail in Chapter 3.)
- Structural proteins of the nucleolus—Since DNA from multiple chromosomes is used to form the nucleolus, it must be taken apart prior to mitosis. Phosphorylation of the structural proteins results in dispersion of the nucleolar proteins and disintegration of the nucleolus.
- A variety of protein kinases that regulate the cytoskeleton—In order for mitosis to happen, the cytoskeleton needs to be completely taken apart and rebuilt. A number of proteins are involved in this, including microtubule-associated proteins (MAPs) and even some actin-binding proteins (ABPs), and must be activated directly or indirectly by the M-CDK.
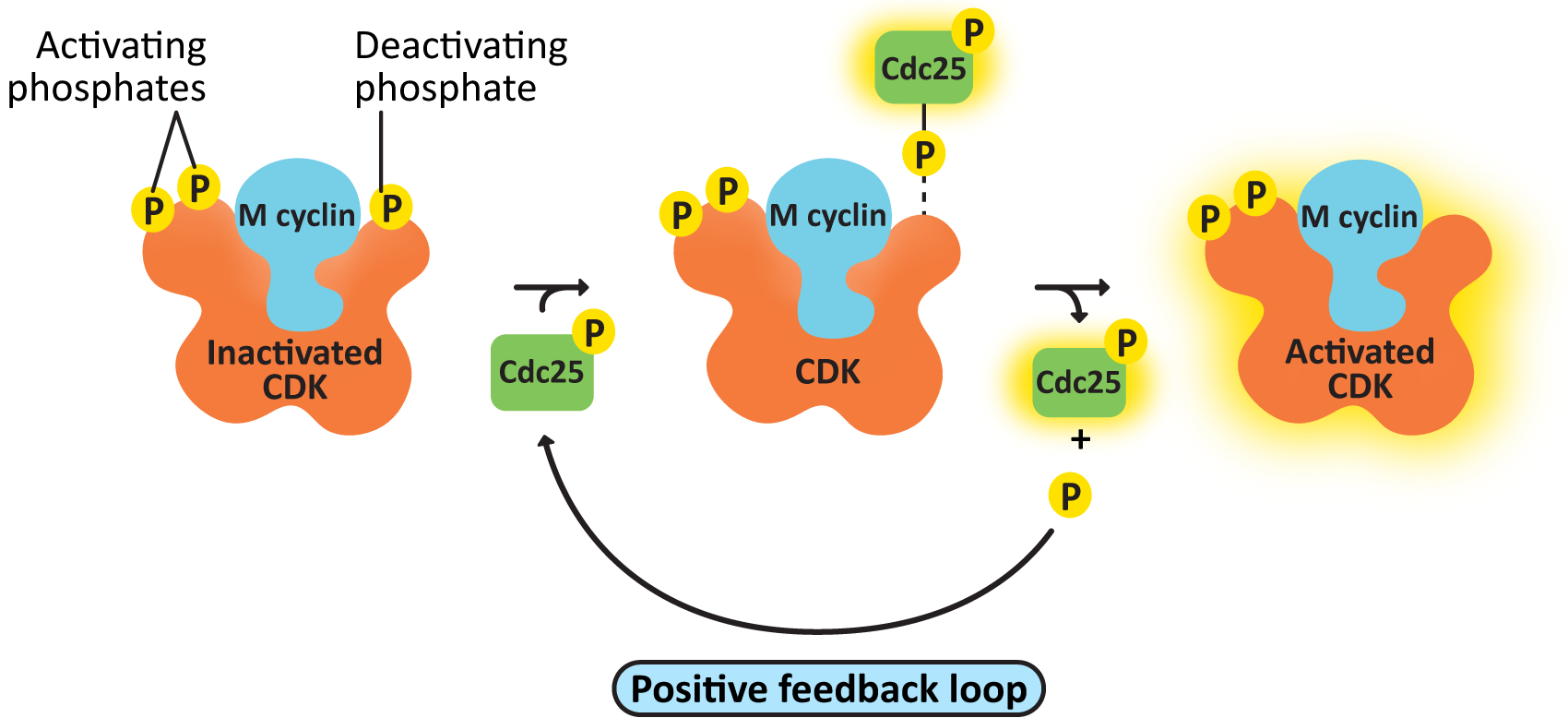
- cdc25, the M-CDK-activating phosphatase—This creates a positive feedback loop, resulting in further activation of M-CDK-cyclin. As a result, M-CDK activity rises increasingly rapidly as more M-CDK becomes active (Figure 08-08).
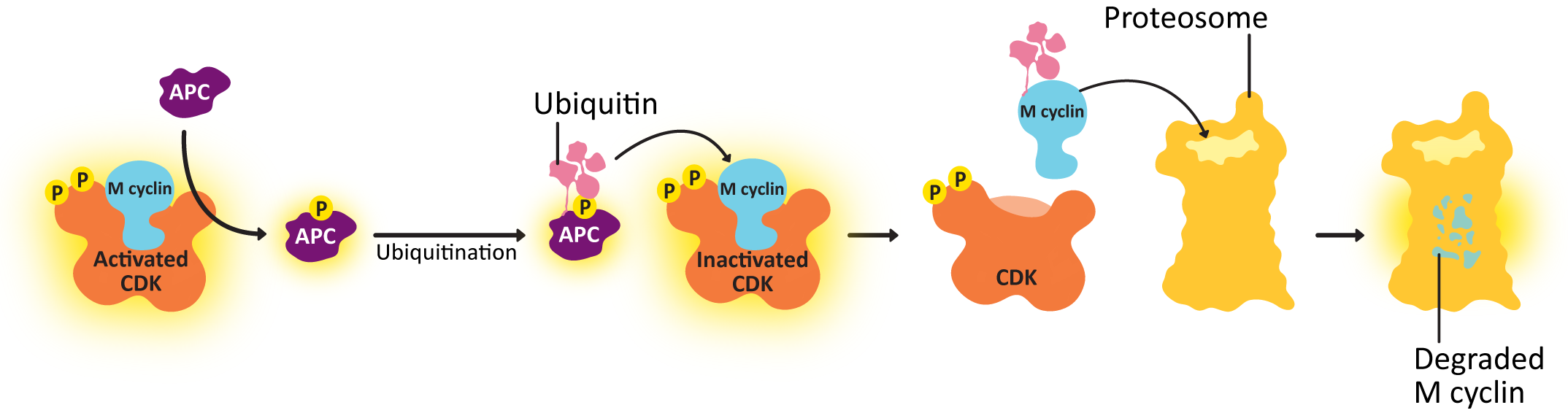
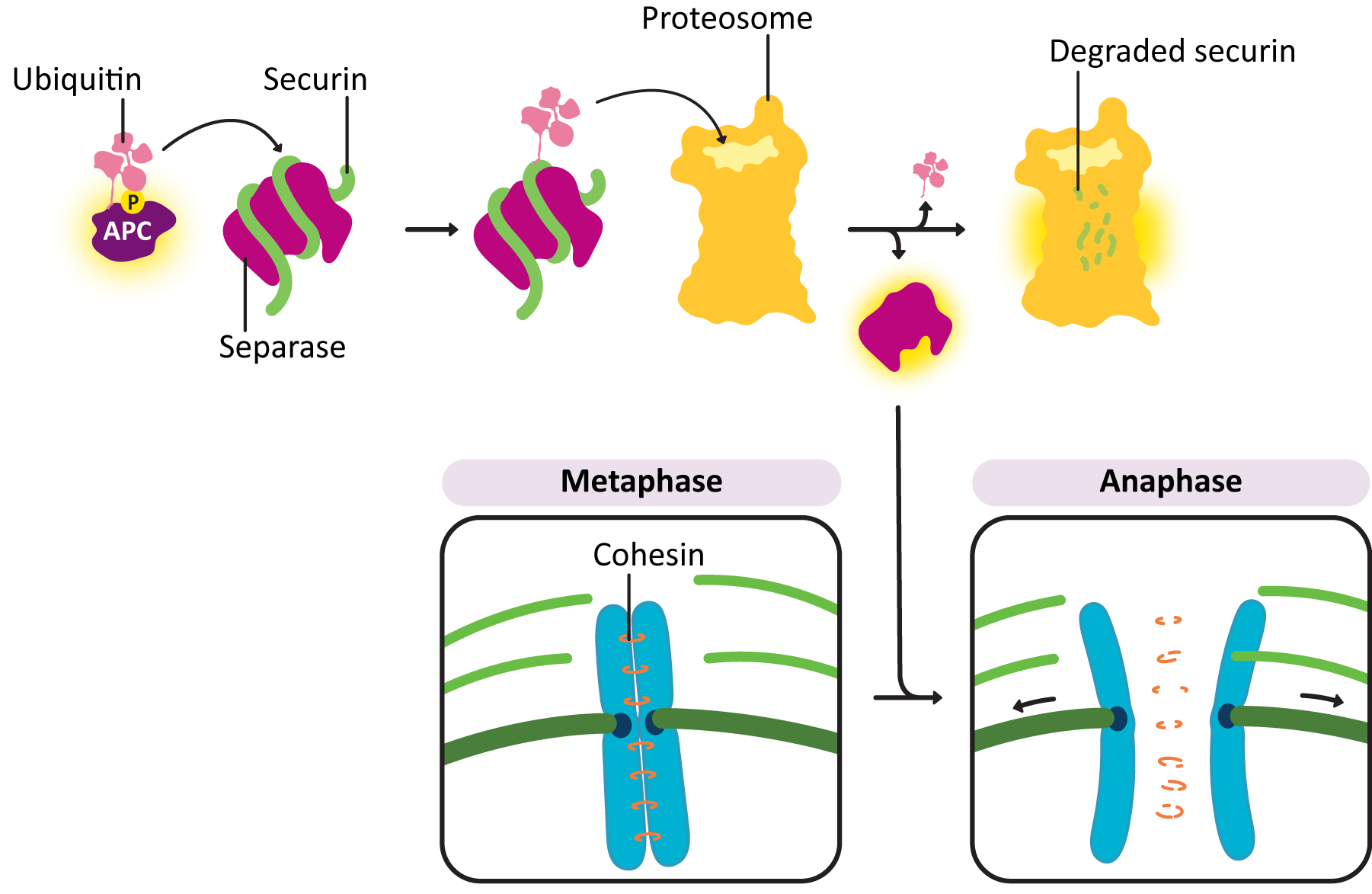
- Anaphase-promoting complex (APC)—This protein is key to passing the metaphase checkpoint. At the beginning of anaphase, APC degrades the cohesin proteins that bind sister chromatids together, releasing the daughter chromosomes. Interestingly, APC also activates enzymes that tag M cyclin for degradation. This ensures that M-CDK will be properly deactivated when its task is done. The cell cannot complete mitosis and return the cytoplasm to its interphase state unless M-CDK is inactive.
Deactivation of the Cyclin-CDK Complex
- shutting down the transcription and translation of new cyclin and
- degrading the cyclin proteins that already exist.
- Chemically synchronizing cells in a population: This technique is the starting point for many experiments to ensure all cells are starting at the same stage in the cell cycle.
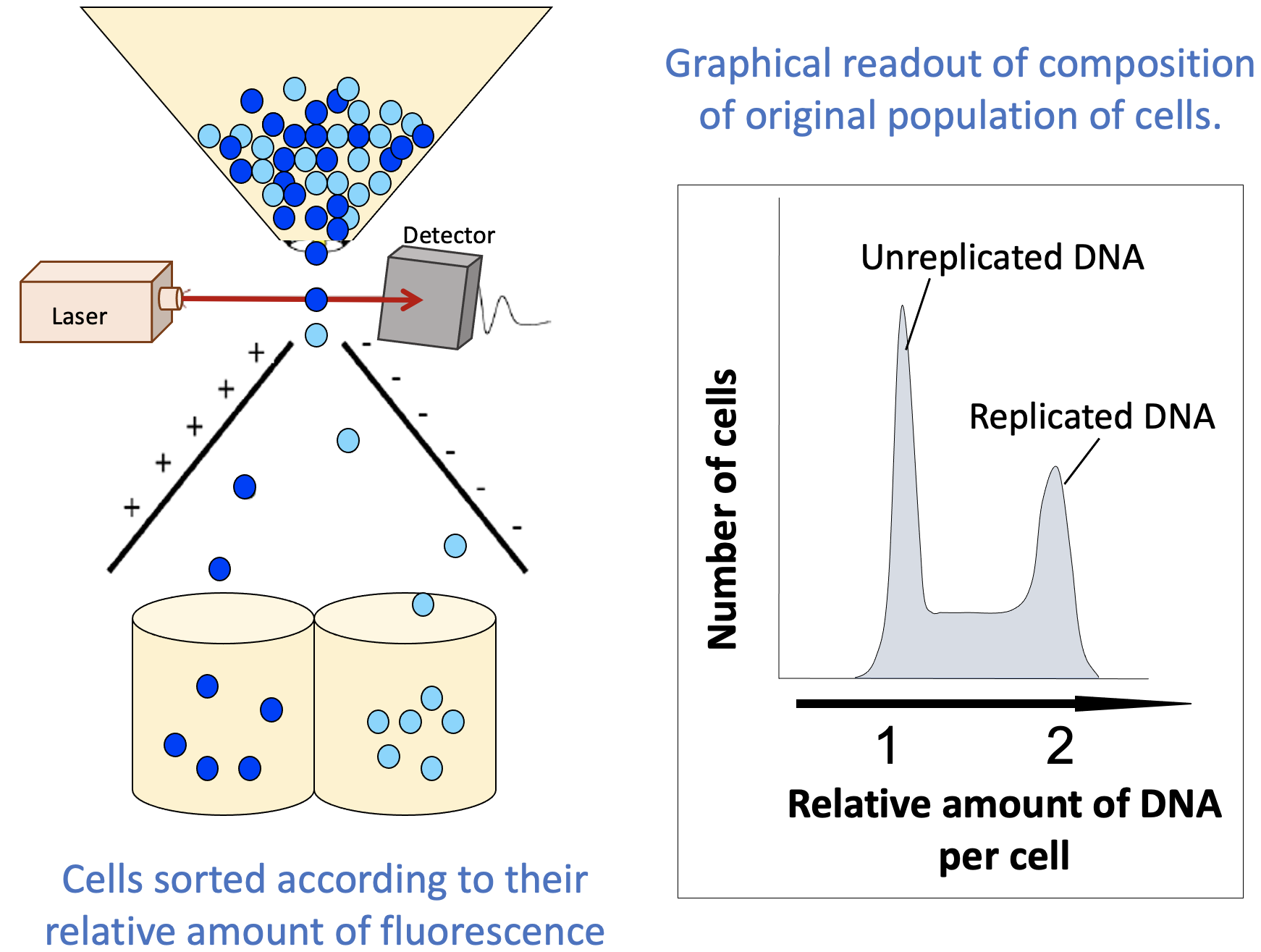
- Fluorescence-activated cell sorting (FACS): This is a subtype of a more commonly known procedure called flow cytometry, where cells are monitored for certain properties and grouped based on these properties.
- In Chapter 4, we discovered the existence of temperature-sensitive mutations, and how they could be used to study the essential proteins of the secretory pathway. Most cell cycle proteins are equally essential, as a cell that cannot progress through its cell cycle cannot divide. So temperature-sensitive mutants are useful in this context as well.
- In Chapter 6, we also saw that chemical inhibitors could be used to disrupt cytoskeletal function temporarily. This, too, is a strategy that can be applied to the cell cycle. There are a few known chemical inhibitors that block further progression in the cell cycle. DNA replication is often targeted by these compounds, and the cell stalls at the start of S phase as a result of the chemical inhibition.
- You now have synchronized populations of cells without using chemical inhibitors or temperature-sensitive mutations. These can be used for further experiments.
- Cells in G1 will have one “full set” of their DNA, while cells in G2 and M phase will have duplicated their DNA (i.e., two “full sets”).
- Cells in S phase will have more than one set but less than two sets, as replication has started in S phase but is not yet complete.
- Additional visual separation may be required to differentiate between G2 and M phase, but this can be done relatively easily, as a cell undergoing mitosis can be identified using a standard light microscope.
- The flow cytometer also produces a graphical readout (Figure 08-10) that summarizes how many cells were found with each “amount” of fluorescent material (DNA in this case). These readouts can be used to learn important information about your population of cells.
- The readout indicates “replicated” and “unreplicated” DNA, not whether it’s in G1, S, G2, or M phase. This is an important limitation of this technique. There are two phases in which the DNA has been replicated and as such are lumped together in the second peak (G2 and M phase). FACS cannot differentiate between these, since the DNA content is the same for both. Depending on the question you’re trying to answer, this might be key to correctly interpreting your results. To separate G2 from M phase in your sample, you would need to do something additional, like look at the cells in a microscope, for example.
- The part in between the peaks is more important than it might seem. In this area, we have more than one “full set” of DNA but less than two. There’s only one stage of the cell cycle where the amount of DNA goes from one set to two: S phase. Cells that are in the process of undergoing S phase will be found between the peaks that represent G1 cells and G2/M cells.
Learning Goals
- Relate the progression of mitosis to the activation and deactivation of proteins by the mitotic CDK-cyclin complex.
- Explain the role of the cytoskeleton (and associated motor proteins) during mitosis and cytokinesis, including how dynamic instability contributes to the formation and function of the mitotic spindle.
- Describe how signaling events are used to end mitosis, starting at anaphase and followed by the transition of the cell back into G1.
- The entire transcription and translation machinery needs to be shut down so that the DNA can be replicated, condensed, accurately divided, and then set up again in the new cell.
- The microtubule network in the cell needs to be dismantled and completely rearranged so that it can help with DNA separation.
- The actin network also needs to be rearranged to help with cytokinesis (division of the cytosol). This means that all of the regular work of the cytoskeleton during interphase (transport, organelle positioning, etc.) is also disrupted.
- Every organelle and structure in the cell must be duplicated and/or distributed into the two daughter cells so that both of the new cells have everything they need to survive. This often requires a complete dismantling of larger organelles (like the endomembrane system or the nucleus).
- A whole series of signaling events needs to be started up to initiate all of this change, and then, at the end, it all needs to be shut down again so that everything can transition back to the interphase and regular cellular function can be rebooted.
Phosphorylation of Histone H1 Promotes Higher-Order Chromatin Packing
Condensin I and Condensin II Work Together to Pack the DNA into a Tight Column
- Mitosis: This is the process of division of the nucleus and its contents (i.e., the DNA!).
- Cytokinesis: This is where the rest of the cytoplasm gets divided into two new cells.
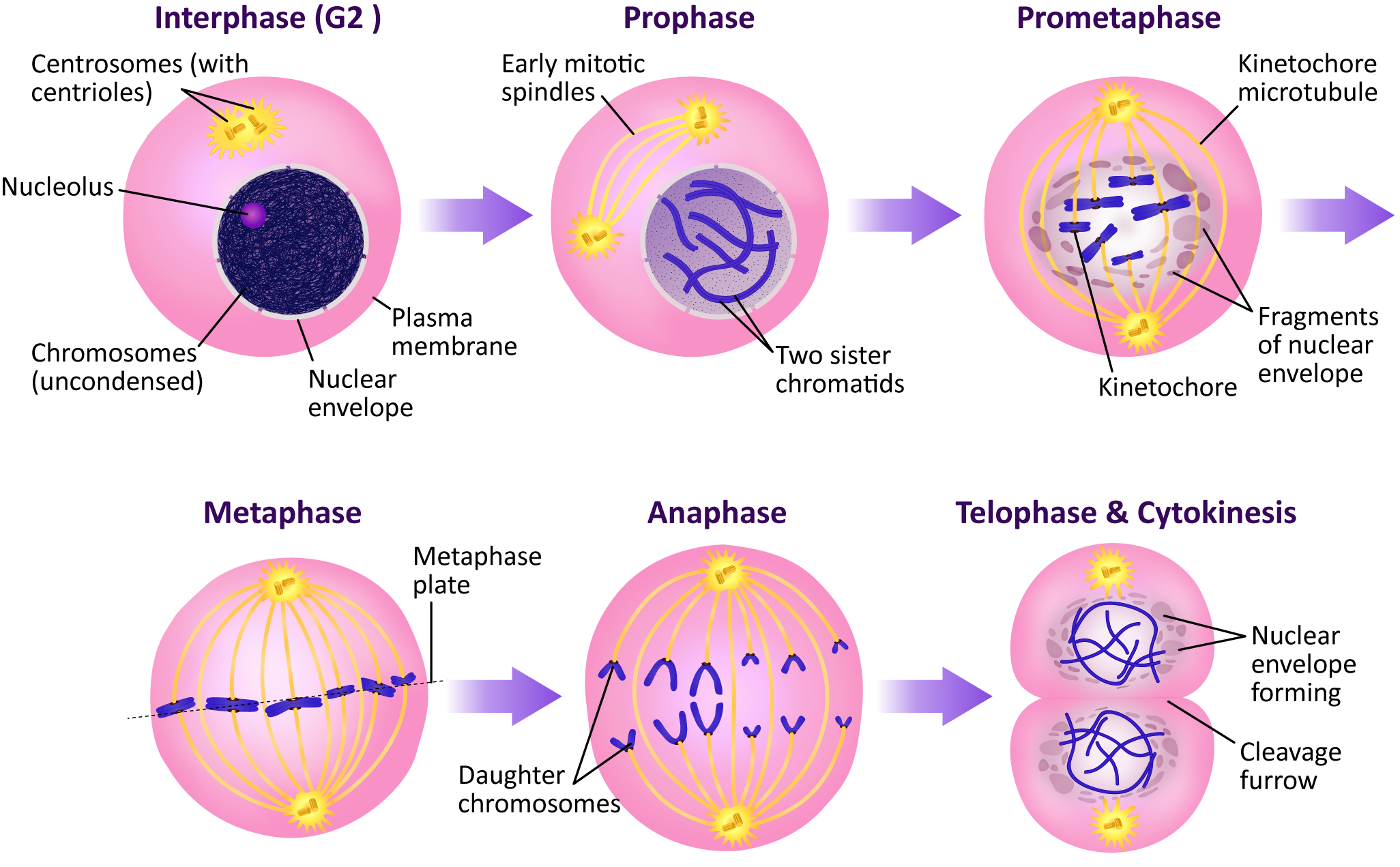
- Chromosomes condense as H1 and nucleolin are phosphorylated.
- The nuclear lamins depolymerize, resulting in the nuclear envelope breaking apart.
- The mitotic spindle will begin to form.
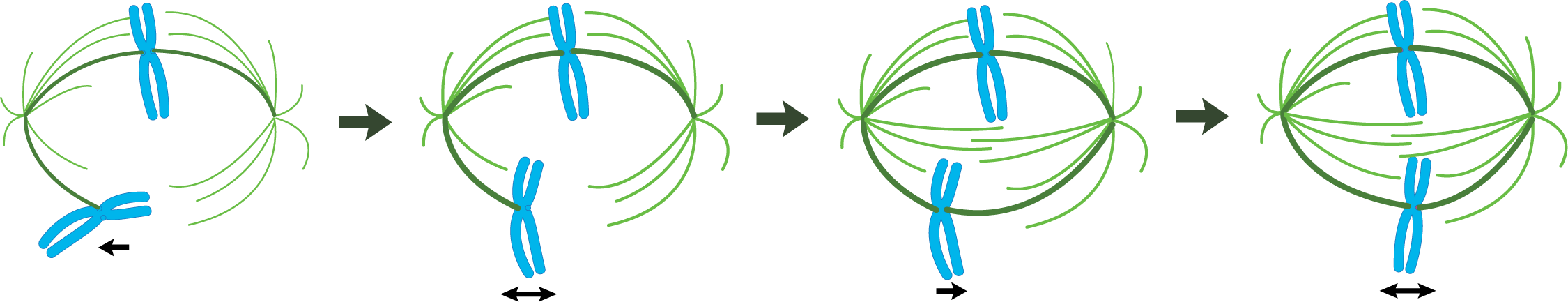
- If microtubules from opposite poles happen to interact with each other, they will stabilize each other (with the help of motor proteins), thus forming a bridge between the two spindle poles. These are aptly named interpolar microtubules, as they span the two poles.
- Conversely, if they happen to interact with a chromosome’s kinetochore during their random growth and shrinkage, they will attach and become a kinetochore microtubule.
- Astral microtubules, the third type of microtubule, are anchored at the plasma membrane (also with the help of motor proteins).
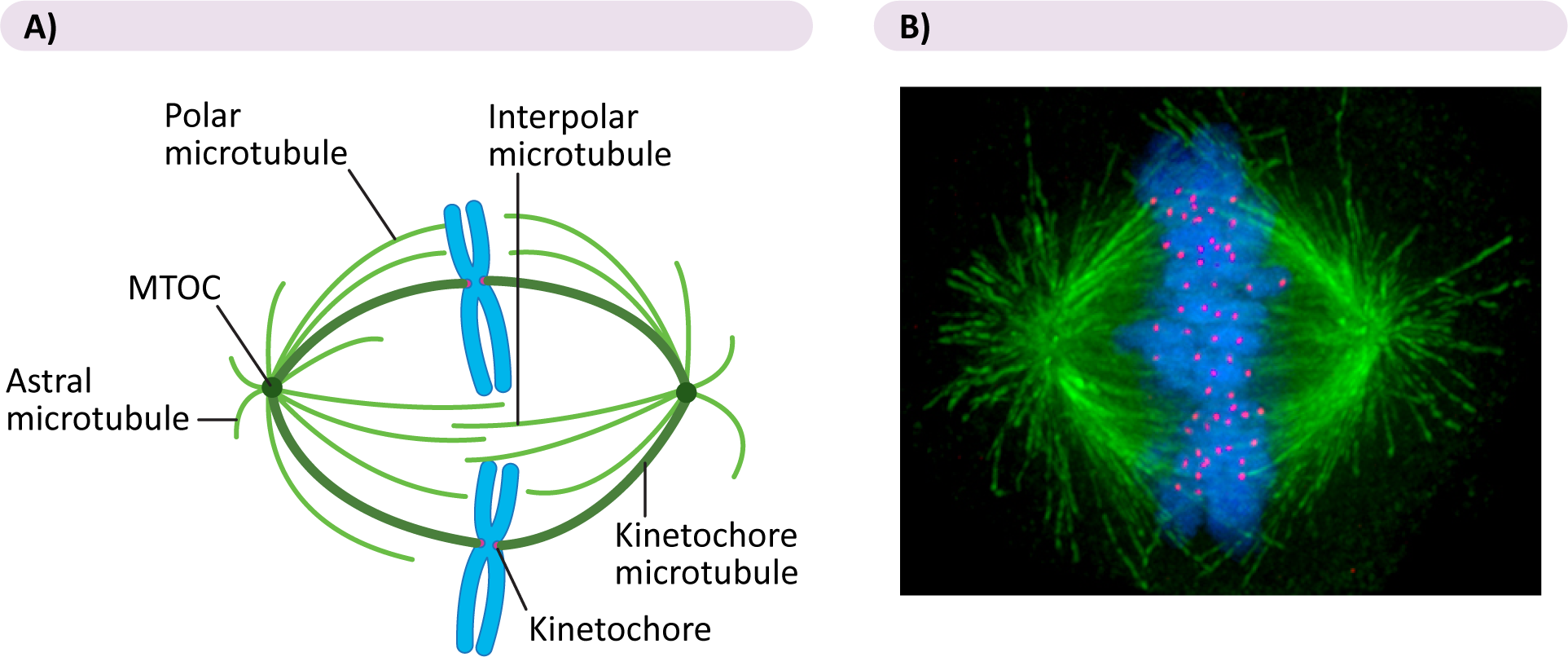
- Kinesins are found at the kinetochore and help the chromosomes stay attached to the kinetochore microtubules as they disassemble in anaphase.
- Dyneins hold the astral microtubules and also help pull the spindle poles apart in anaphase
- Interpolar microtubules have a special double-headed kinesin, which walks along each of the interpolar microtubules in a set. They also help to push the spindle poles apart in anaphase B.
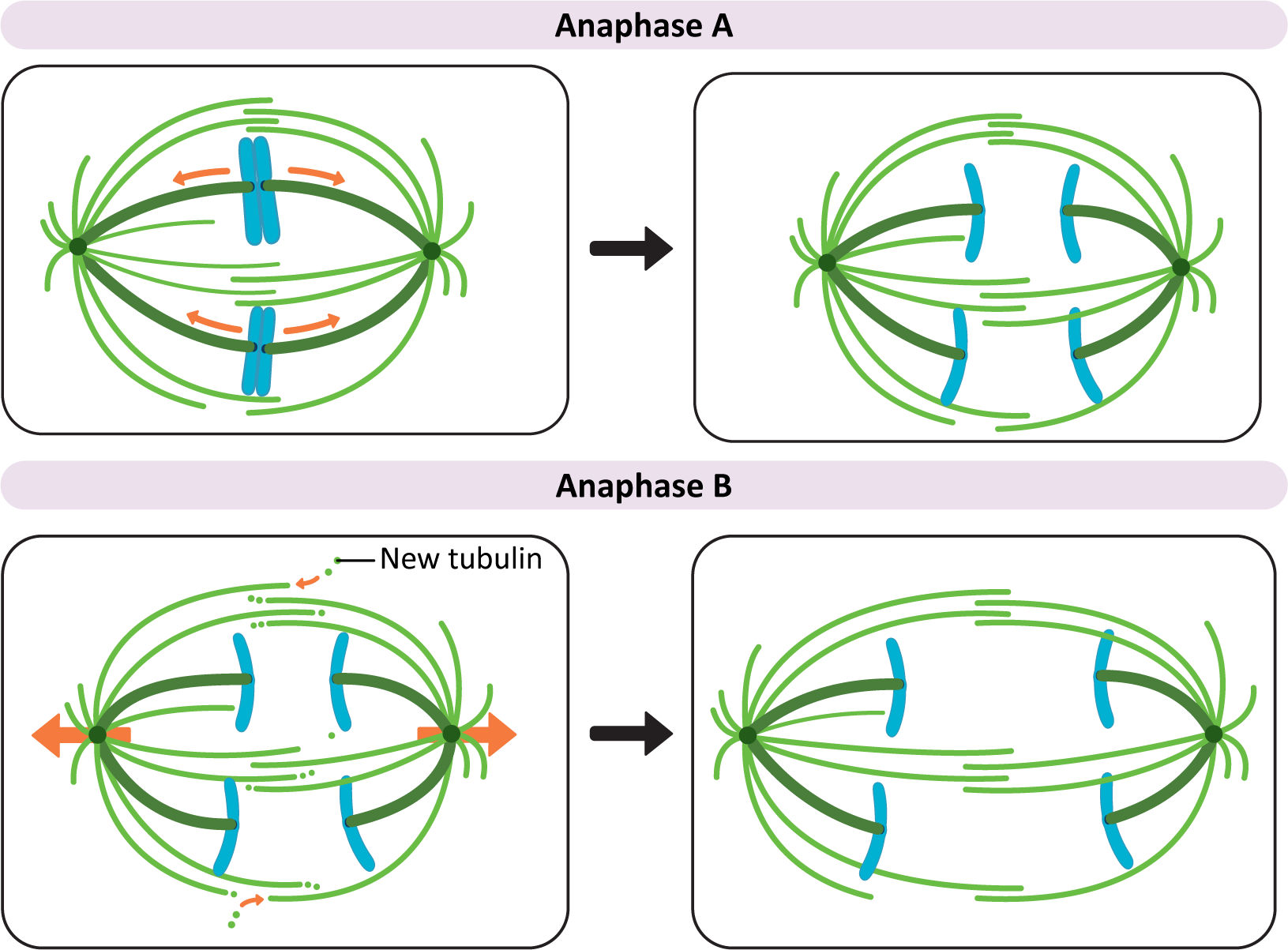
- Anaphase A. During the initial part of anaphase, kinetochore microtubules shorten and move the chromosomes toward the opposite poles. This requires a combination of depolymerization of kinetochore microtubules and the action of motor proteins at the kinetochore that help keep the chromosome connected to the shrinking microtubule.
- Anaphase B. During anaphase B, the length of the kinetochore microtubules (MTs) remains more or less constant. Now it is the other MTs of the mitotic spindle that begin to do the work. Again, there is much that is still unknown about this process, but three separate forces have been shown to play a role in anaphase B:
- Tubulin subunits are added to the plus ends of the interpolar microtubules, making them longer.
- Motor proteins move the overlapping interphase microtubules from the two poles apart to make the spindle longer. This requires the help of the kinesins that are holding the paired interpolar microtubules together.
- Dyneins, attached to the astral microtubules and the cell cortex, walk toward the spindle poles on either side. This results in a shortening of the distance between the poles of the spindle and the plasma membrane. It also helps pull the spindle poles farther apart from each other.
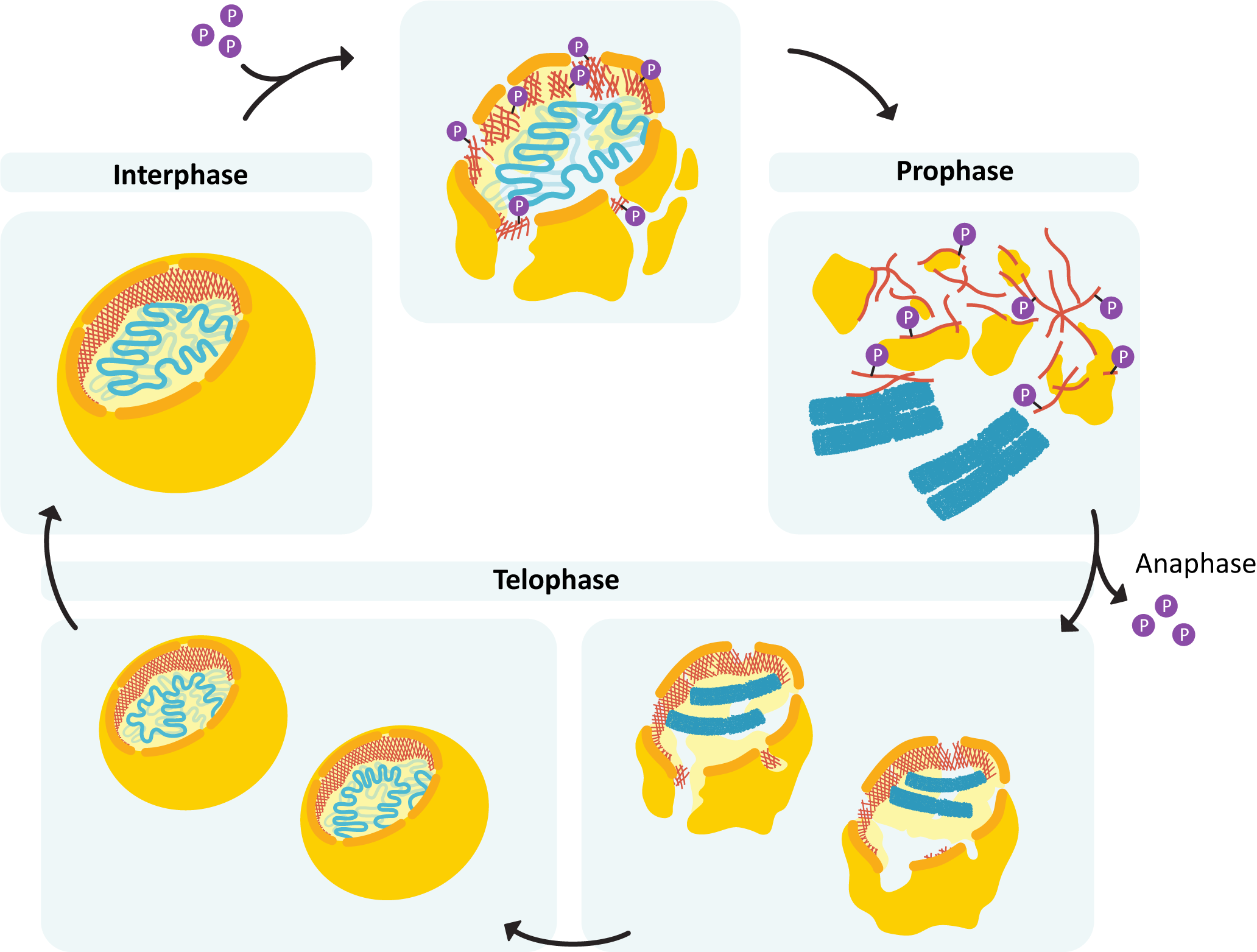
- The nuclear lamina must reassemble so that the nuclear envelope can be rebuilt around the chromosome.
- The mitotic spindle disintegrates, and the cytoskeleton reassembles into its interphase conformation.
- The chromosomes must decondense and reform their interphase arrangement, which includes the reassembly of the nucleolus.
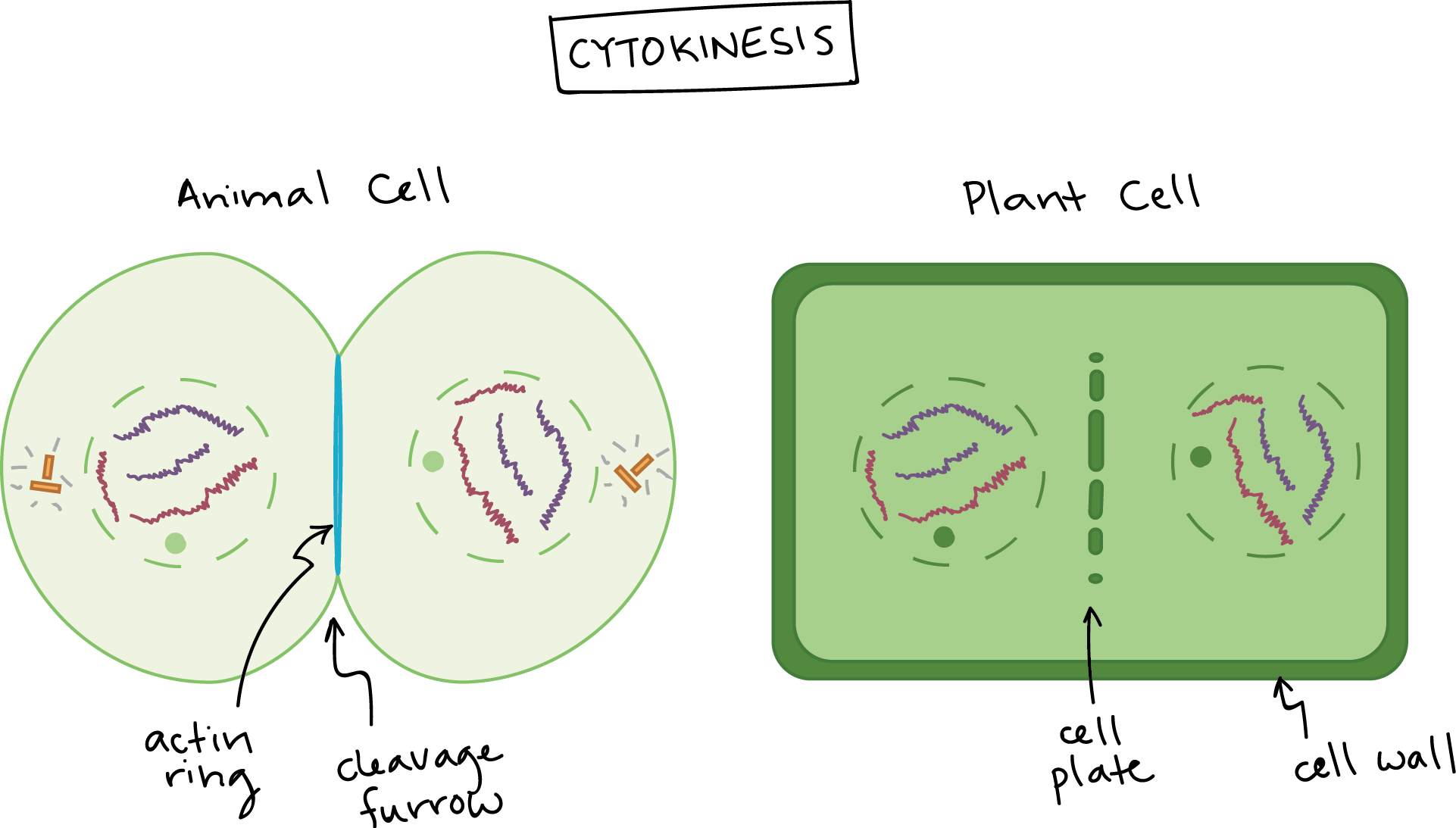
Cytokinesis in Plant Cells
Mitosis and Cytokinesis in Other Eukaryotes
- Plant and animal cells undergo what’s called open mitosis, which means that the nuclear envelope breaks down completely in order to form the mitotic spindle. There are also two other forms:
- Partially closed mitosis occurs when the nuclear envelope remains intact and the spindle is formed both inside and outside of the nuclear envelope. Budding yeast (Saccharomyces cerevisiae) uses a partially closed format, which is especially interesting considering how often they are used as a model organism for all Eukaryotes.
- A third form is closed mitosis, which happens when the nuclear envelope not only remains intact, but the nuclear pores are plugged to close the nucleus off completely from the rest of the cell. This is more common in protists, fungi, and other single-celled organisms.
- Many cell types don’t divide evenly at mitosis. Lots of plant and animal cells undergo asymmetric division, which results in one daughter cell that is larger than the other. Additionally, lots of cells “grow” their daughter cell off of one side and move the new daughter nuclei into the bud once the bud is large enough. Saccharomyces cerevisiae is called “budding yeast” for this very reason. There are many additional cells that do this, especially single-celled algae, fungi, and other protists.
- Signaling: The cell cycle requires integrated information from the extracellular and intracellular environments through signaling. The balance of activating forces versus inactivating forces helps ensure that cells divide only when the time is right.
- Cytoskeleton: The cytoskeleton is highly involved, especially in M phase. It’s clear that dynamic instability, a hallmark of the cytoskeletal system, is vital for the formation of the mitotic spindle but also the correct alignment of the chromosomes at the metaphase plate.
- DNA packing: The molecular packers and organization help unpack all the DNA during synthesis and do the opposite in M phase, completely packaging the DNA.
- Other organelles: Of course, these organelles need to be properly divided between daughter cells, but that does not end their contribution to this process. Prior to shutdown for mitosis, they need to make sure that everything the cell will need is available. This will include a great deal of energy, in the form of ATP and GTP, membrane, vesicles, and even polysaccharides if a wall needs to be made between the two daughter cells.
Topic 8.1: Regulating the Cell Cycle: Checkpoint Control
- Make a list of the four phases of the cell cycle, and describe in detail what has to happen in each phase before it is complete. Be as specific as you can.
- For each of the stages, make a list of experimental techniques that could be used to identify cells that are in each of the four stages. Explain the rationale for your choice in each case.
- Explain the concept of checkpoint control of the cell cycle and give examples of what types of conditions should be met before S and M phases proceed. Explain why those conditions need to be met before the cell moves to the next phase.
- Describe the sequence of events leading to CDK activation and deactivation.
- Define negative and positive feedback. How are they thought to play a role in cell cycle control?
- Discuss the role of CDK-cyclin complexes in regulation of the cell cycle. How do they contribute to the cell “knowing” the conditions are right for the next stage of the cell cycle?
- Compare and contrast the roles of CDKs that control entry into S phase versus entry into M phase. Make a list of the different examples of proteins that they are thought to interact with and their effects.
- Explain the role of phosphorylation in regulation of the cell cycle. Explain how phosphorylation can affect protein folding and how that might result in changes in protein activity.
- Describe the role of dynamic instability in the formation and maintenance of the mitotic spindle.
- Illustrate with labeled diagrams the relationship between chromatids and chromosomes at each of the stages of the cell cycle.
- Explain how each stage of mitosis is ultimately controlled by the activation and deactivation of CDKs.
- Compare and contrast the following stages:
- prophase and prometaphase
- prophase and telophase
- anaphase A and B
- What is cytokinesis, and how does it differ in plants and animals?

Comments
Post a Comment